Find the Exact Solution for the Beverton Holt Model
Determining The Optimal Number Of Clusters: 3 Must Know Methods
Determining the optimal number of clusters in a data set is a fundamental issue in partitioning clustering, such as k-means clustering, which requires the user to specify the number of clusters k to be generated.
Unfortunately, there is no definitive answer to this question. The optimal number of clusters is somehow subjective and depends on the method used for measuring similarities and the parameters used for partitioning.
A simple and popular solution consists of inspecting the dendrogram produced using hierarchical clustering to see if it suggests a particular number of clusters. Unfortunately, this approach is also subjective.
In this chapter, we'll describe different methods for determining the optimal number of clusters for k-means, k-medoids (PAM) and hierarchical clustering.
These methods include direct methods and statistical testing methods:
- Direct methods: consists of optimizing a criterion, such as the within cluster sums of squares or the average silhouette. The corresponding methods are named elbow and silhouette methods, respectively.
- Statistical testing methods: consists of comparing evidence against null hypothesis. An example is the gap statistic.
In addition to elbow, silhouette and gap statistic methods, there are more than thirty other indices and methods that have been published for identifying the optimal number of clusters. We'll provide R codes for computing all these 30 indices in order to decide the best number of clusters using the "majority rule".
For each of these methods:
- We'll describe the basic idea and the algorithm
- We'll provide easy-o-use R codes with many examples for determining the optimal number of clusters and visualizing the output.
Contents:
- Elbow method
- Average silhouette method
- Gap statistic method
- Computing the number of clusters using R
- Required R packages
- Data preparation
- fviz_nbclust() function: Elbow, Silhouhette and Gap statistic methods
- NbClust() function: 30 indices for choosing the best number of clusters
- Summary
- References
Related Book
Practical Guide to Cluster Analysis in R
Elbow method
Recall that, the basic idea behind partitioning methods, such as k-means clustering, is to define clusters such that the total intra-cluster variation [or total within-cluster sum of square (WSS)] is minimized. The total WSS measures the compactness of the clustering and we want it to be as small as possible.
The Elbow method looks at the total WSS as a function of the number of clusters: One should choose a number of clusters so that adding another cluster doesn't improve much better the total WSS.
The optimal number of clusters can be defined as follow:
- Compute clustering algorithm (e.g., k-means clustering) for different values of k. For instance, by varying k from 1 to 10 clusters.
- For each k, calculate the total within-cluster sum of square (wss).
- Plot the curve of wss according to the number of clusters k.
- The location of a bend (knee) in the plot is generally considered as an indicator of the appropriate number of clusters.
Note that, the elbow method is sometimes ambiguous. An alternative is the average silhouette method (Kaufman and Rousseeuw [1990]) which can be also used with any clustering approach.
Average silhouette method
The average silhouette approach we'll be described comprehensively in the chapter cluster validation statistics. Briefly, it measures the quality of a clustering. That is, it determines how well each object lies within its cluster. A high average silhouette width indicates a good clustering.
Average silhouette method computes the average silhouette of observations for different values of k. The optimal number of clusters k is the one that maximize the average silhouette over a range of possible values for k (Kaufman and Rousseeuw 1990).
The algorithm is similar to the elbow method and can be computed as follow:
- Compute clustering algorithm (e.g., k-means clustering) for different values of k. For instance, by varying k from 1 to 10 clusters.
- For each k, calculate the average silhouette of observations ( a v g.s i l ).
- Plot the curve of a v g.s i l according to the number of clusters k.
- The location of the maximum is considered as the appropriate number of clusters.
Gap statistic method
The gap statistic has been published by R. Tibshirani, G. Walther, and T. Hastie (Standford University, 2001). The approach can be applied to any clustering method.
The gap statistic compares the total within intra-cluster variation for different values of k with their expected values under null reference distribution of the data. The estimate of the optimal clusters will be value that maximize the gap statistic (i.e, that yields the largest gap statistic). This means that the clustering structure is far away from the random uniform distribution of points.
The algorithm works as follow:
- Cluster the observed data, varying the number of clusters from k = 1, …, k m a x , and compute the corresponding total within intra-cluster variation W k .
- Generate B reference data sets with a random uniform distribution. Cluster each of these reference data sets with varying number of clusters k = 1, …, k m a x , and compute the corresponding total within intra-cluster variation W k b .
- Compute the estimated gap statistic as the deviation of the observed W k value from its expected value W k b under the null hypothesis: \(Gap(k) = \frac{1}{B} \sum\limits_{b=1}^B log(W_{kb}^*) - log(W_k)\) . Compute also the standard deviation of the statistics.
- Choose the number of clusters as the smallest value of k such that the gap statistic is within one standard deviation of the gap at k+1: G a p(k)≥G a p(k + 1)−s k + 1 .
Note that, using B = 500 gives quite precise results so that the gap plot is basically unchanged after an another run.
Computing the number of clusters using R
In this section, we'll describe two functions for determining the optimal number of clusters:
- fviz_nbclust() function [in factoextra R package]: It can be used to compute the three different methods [elbow, silhouette and gap statistic] for any partitioning clustering methods [K-means, K-medoids (PAM), CLARA, HCUT]. Note that the hcut() function is available only in factoextra package.It computes hierarchical clustering and cut the tree in k pre-specified clusters.
- NbClust() function [ in NbClust R package] (Charrad et al. 2014): It provides 30 indices for determining the relevant number of clusters and proposes to users the best clustering scheme from the different results obtained by varying all combinations of number of clusters, distance measures, and clustering methods. It can simultaneously computes all the indices and determine the number of clusters in a single function call.
Required R packages
We'll use the following R packages:
- factoextra to determine the optimal number clusters for a given clustering methods and for data visualization.
- NbClust for computing about 30 methods at once, in order to find the optimal number of clusters.
To install the packages, type this:
pkgs <- c("factoextra", "NbClust") install.packages(pkgs) Load the packages as follow:
library(factoextra) library(NbClust) Data preparation
We'll use the USArrests data as a demo data set. We start by standardizing the data to make variables comparable.
# Standardize the data df <- scale(USArrests) head(df) ## Murder Assault UrbanPop Rape ## Alabama 1.2426 0.783 -0.521 -0.00342 ## Alaska 0.5079 1.107 -1.212 2.48420 ## Arizona 0.0716 1.479 0.999 1.04288 ## Arkansas 0.2323 0.231 -1.074 -0.18492 ## California 0.2783 1.263 1.759 2.06782 ## Colorado 0.0257 0.399 0.861 1.86497 fviz_nbclust() function: Elbow, Silhouhette and Gap statistic methods
The simplified format is as follow:
fviz_nbclust(x, FUNcluster, method = c("silhouette", "wss", "gap_stat")) - x: numeric matrix or data frame
- FUNcluster: a partitioning function. Allowed values include kmeans, pam, clara and hcut (for hierarchical clustering).
- method: the method to be used for determining the optimal number of clusters.
The R code below determine the optimal number of clusters for k-means clustering:
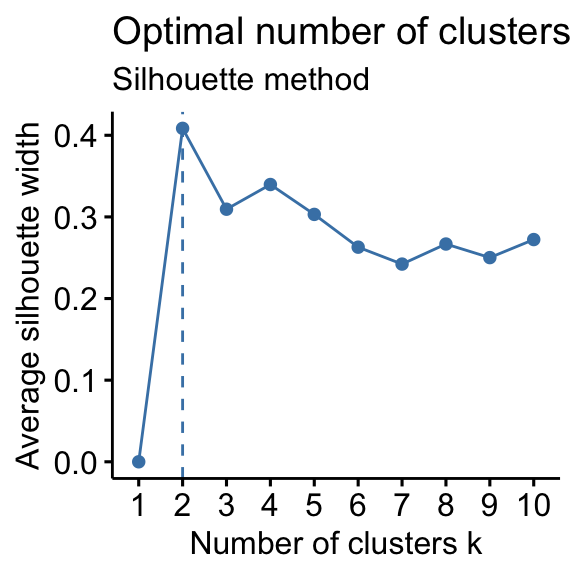
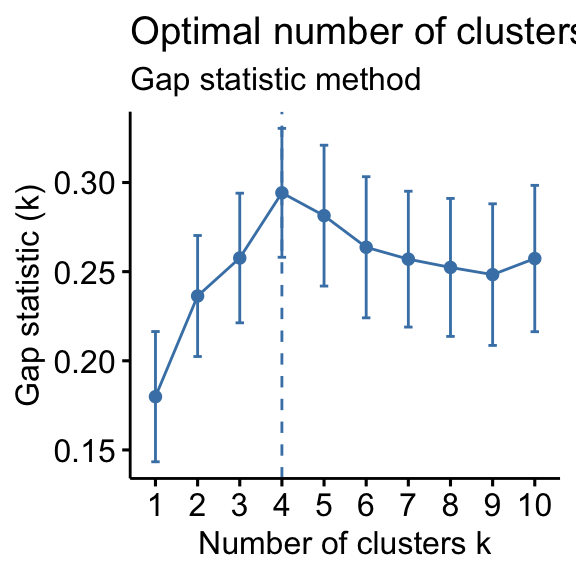
# Elbow method fviz_nbclust(df, kmeans, method = "wss") + geom_vline(xintercept = 4, linetype = 2)+ labs(subtitle = "Elbow method") # Silhouette method fviz_nbclust(df, kmeans, method = "silhouette")+ labs(subtitle = "Silhouette method") # Gap statistic # nboot = 50 to keep the function speedy. # recommended value: nboot= 500 for your analysis. # Use verbose = FALSE to hide computing progression. set.seed(123) fviz_nbclust(df, kmeans, nstart = 25, method = "gap_stat", nboot = 50)+ labs(subtitle = "Gap statistic method") ## Clustering k = 1,2,..., K.max (= 10): .. done ## Bootstrapping, b = 1,2,..., B (= 50) [one "." per sample]: ## .................................................. 50 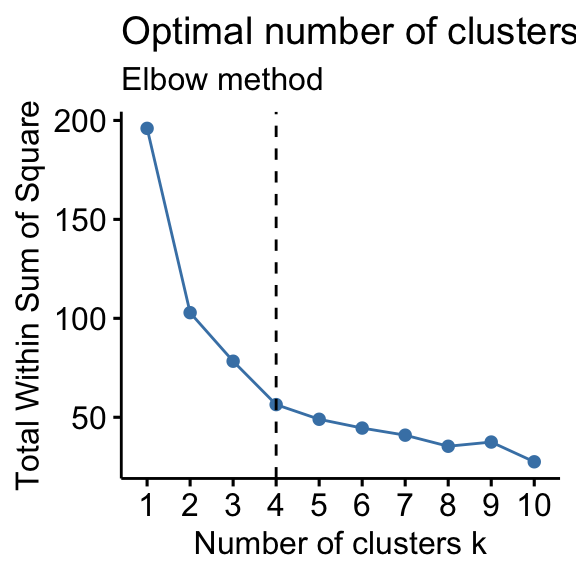
- Elbow method: 4 clusters solution suggested
- Silhouette method: 2 clusters solution suggested
- Gap statistic method: 4 clusters solution suggested
According to these observations, it's possible to define k = 4 as the optimal number of clusters in the data.
The disadvantage of elbow and average silhouette methods is that, they measure a global clustering characteristic only. A more sophisticated method is to use the gap statistic which provides a statistical procedure to formalize the elbow/silhouette heuristic in order to estimate the optimal number of clusters.
NbClust() function: 30 indices for choosing the best number of clusters
The simplified format of the function NbClust() is:
NbClust(data = NULL, diss = NULL, distance = "euclidean", min.nc = 2, max.nc = 15, method = NULL) - data: matrix
- diss: dissimilarity matrix to be used. By default, diss=NULL, but if it is replaced by a dissimilarity matrix, distance should be "NULL"
- distance: the distance measure to be used to compute the dissimilarity matrix. Possible values include "euclidean", "manhattan" or "NULL".
- min.nc, max.nc: minimal and maximal number of clusters, respectively
- method: The cluster analysis method to be used including "ward.D", "ward.D2", "single", "complete", "average", "kmeans" and more.
- To compute NbClust() for kmeans, use method = "kmeans".
- To compute NbClust() for hierarchical clustering, method should be one of c("ward.D", "ward.D2", "single", "complete", "average").
The R code below computes NbClust() for k-means:
Here, there are contents hidden to non-premium members. Sign up now to read all of our premium contents and to be awarded a certificate of course completion.
Claim Your Membership Now
## Among all indices: ## =================== ## * 2 proposed 0 as the best number of clusters ## * 10 proposed 2 as the best number of clusters ## * 2 proposed 3 as the best number of clusters ## * 8 proposed 4 as the best number of clusters ## * 1 proposed 5 as the best number of clusters ## * 1 proposed 8 as the best number of clusters ## * 2 proposed 10 as the best number of clusters ## ## Conclusion ## ========================= ## * According to the majority rule, the best number of clusters is 2 . 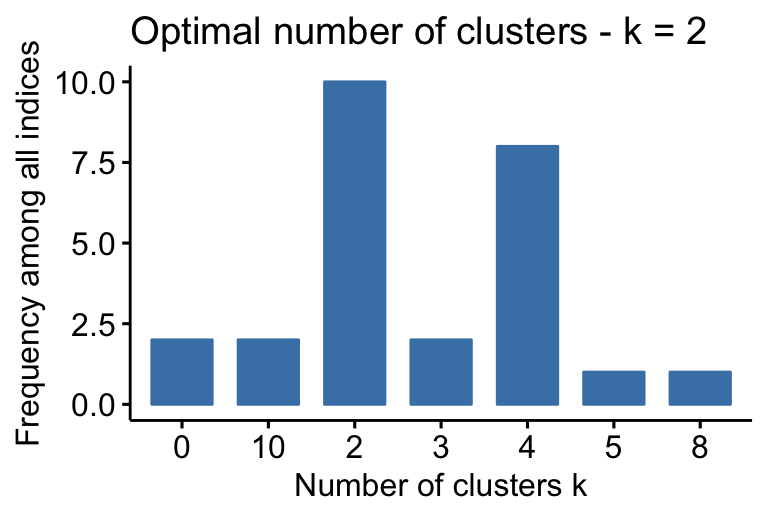
- ….
- 2 proposed 0 as the best number of clusters
- 10 indices proposed 2 as the best number of clusters.
- 2 proposed 3 as the best number of clusters.
- 8 proposed 4 as the best number of clusters.
According to the majority rule, the best number of clusters is 2.
Summary
In this article, we described different methods for choosing the optimal number of clusters in a data set. These methods include the elbow, the silhouette and the gap statistic methods.
We demonstrated how to compute these methods using the R function fviz_nbclust() [in factoextra R package]. Additionally, we described the package NbClust(), which can be used to compute simultaneously many other indices and methods for determining the number of clusters.
After choosing the number of clusters k, the next step is to perform partitioning clustering as described at: k-means clustering.
References
Charrad, Malika, Nadia Ghazzali, Véronique Boiteau, and Azam Niknafs. 2014. "NbClust: An R Package for Determining the Relevant Number of Clusters in a Data Set." Journal of Statistical Software 61: 1–36. http://www.jstatsoft.org/v61/i06/paper.
Kaufman, Leonard, and Peter Rousseeuw. 1990. Finding Groups in Data: An Introduction to Cluster Analysis.
Recommended for you
This section contains best data science and self-development resources to help you on your path.
Teacher
Find the Exact Solution for the Beverton Holt Model
Source: https://www.datanovia.com/en/lessons/determining-the-optimal-number-of-clusters-3-must-know-methods/